Tutorial 1: Continuum Imaging#
This script was written for CASA 6.6.4.34
Source: First Look at Imaging (CASA Guide)
https://casaguides.nrao.edu/index.php/First_Look_at_Imaging_CASA_6.5.4
Adapted for ALMA Data Processing Workshop @ University of Victoria
J. Speedie (jspeedie@uvic.ca) October 2024
Dataset: TW Hya calibrated continuum measurement set (435 MB)
twhya_calibrated.ms.tar (Download)
Your working directory should look like:
TUTORIAL1 >> ls
script_continuum_imaging.py
twhya_calibrated.ms.tar
Untar with:
tar -xvzf twhya_calibrated.ms.tar
Getting oriented with the data#
listobs(vis='twhya_calibrated.ms',
listfile='twhya_calibrated_listobs.txt')
Inspecting the data#
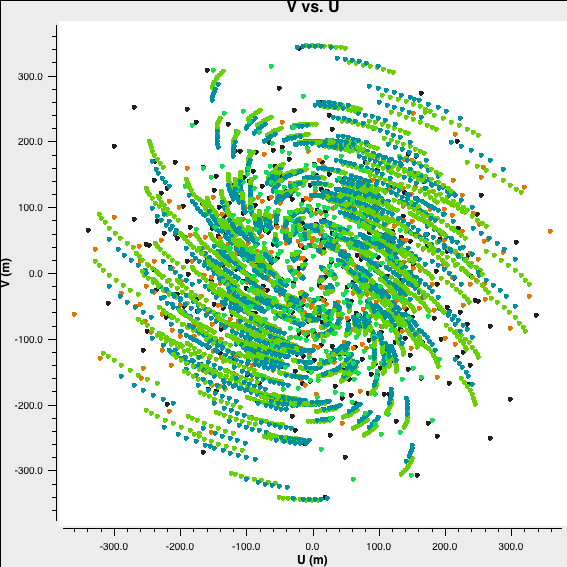
Check the uv coverage:
plotms(vis='twhya_calibrated.ms',
xaxis='u',
yaxis='v',
avgchannel='10000',
avgspw=False,
avgtime='1e9',
avgscan=False,
coloraxis='field',
showgui=True)
Save resulting plot as png file directly:
plotms(vis='twhya_calibrated.ms',
xaxis='u',
yaxis='v',
avgchannel='10000',
avgspw=False,
avgtime='1e9',
avgscan=False,
coloraxis='field',
showgui=False,
plotfile='twhya_calibrated.ms_uvcoverage.png')
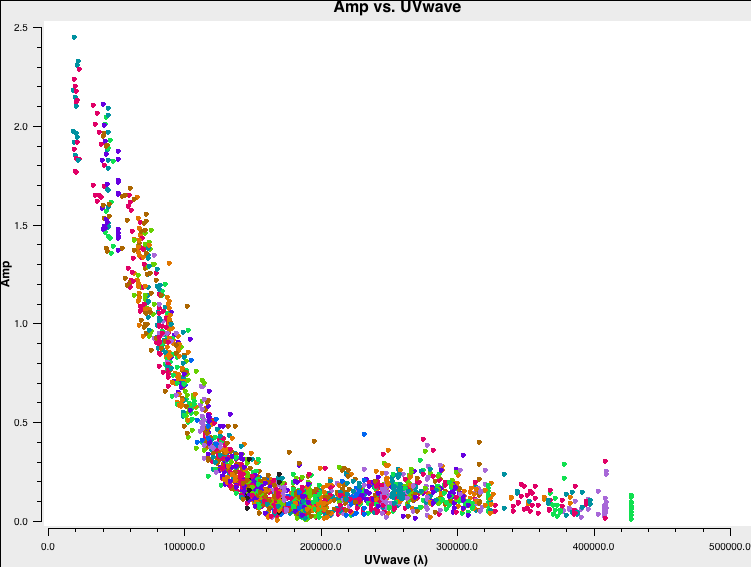
Check the baseline coverage:
plotms(vis='twhya_calibrated.ms',
xaxis='UVwave',
yaxis='Amp',
avgchannel='10000',
avgspw=False,
avgtime='1e9',
avgscan=False,
field='5', # TW Hya
coloraxis='antenna1',
showgui=True)
Save resulting plot as png file directly:
plotms(vis='twhya_calibrated.ms',
xaxis='UVwave',
yaxis='Amp',
avgchannel='10000',
avgspw=False,
avgtime='1e9',
avgscan=False,
field='5', # TW Hya
coloraxis='antenna1',
showgui=False,
plotfile='twhya_calibrated.ms_baselinecoverage.png')
Split out the science target#
os.system('rm -rf twhya_smoothed.ms') # We're about to create this
split(vis='twhya_calibrated.ms', # Old measurement set (435 MB)
field='5', # TW Hya
width='8',
outputvis='twhya_smoothed.ms', # New measurement set (43 MB)
datacolumn='data')
Inspect the new MS:
listobs(vis='twhya_smoothed.ms',
listfile='twhya_smoothed_listobs.txt')
Image the science target#
os.system('rm -rf twhya_cont.*') # We're about to create these
To auto-mask and image non-interactively:
tclean(vis='twhya_smoothed.ms',
imagename='twhya_cont',
field='0',
spw='',
specmode='mfs',
gridder='standard',
deconvolver='hogbom',
imsize=[250,250],
cell=['0.1arcsec'],
weighting='briggs',
robust=0.5,
threshold='0mJy',
niter=5000,
interactive=False,
nmajor = 1, # runs 1 major cycle and stops
usemask='auto-multithresh', # Automasking parameters below this line
noisethreshold=4.25,
sidelobethreshold=2.0,
lownoisethreshold=1.5,
minbeamfrac=0.3,
negativethreshold=15.0,
verbose=True,
fastnoise=False)
Alternative: Image interactively and mask manually in the GUI
tclean(vis='twhya_smoothed.ms',
imagename='twhya_cont',
field='0',
spw='',
specmode='mfs',
gridder='standard',
deconvolver='hogbom',
imsize=[250,250],
cell=['0.1arcsec'],
weighting='briggs',
robust=0.5,
threshold='0mJy',
niter=5000,
interactive=True)
Open & inspect resulting twhya_cont* files in CARTA!
Perform primary beam correction#
os.system('rm -rf twhya_cont.pbcor.image')
Perform primary beam correction and generate primary beam corrected image:
impbcor(imagename='twhya_cont.image',
pbimage='twhya_cont.pb',
outfile='twhya_cont.pbcor.image')